Computational modeling tools and hands-on exercises developed for:
- Math model simulations demo
- Tumor Organoid Model (NIH-PSOP)
- Drugs Interstitial Transport Model (Moffitt PhD Program)
- Targeted Therapy Model (ACS-IRG & NIH-PSOP)
- Drug Resistance Model (WhAM)
- Drug Scheduling Model (HIP-IMO, PASI, Pomona)
- Clonogenic Assay Model (NIH-ICBP, M4M)
- Tumor Growth Model (MBI)
- Deformable Cell Model "IBCell", the Immersed Boundary Model (IAS & Moffitt PhD Program)
- Drug Penetration Model (PASI, PSOC)
Demo movies from a collection of our models
Demo simulations from a collection of our computational models [click here] Created by K.Rejniak
Tumor Organoid Model (NIH PSOP grants)
This project is supported by the NIH- PSOP ~ Physical Sciences Oncology Project grant.
Created by A. Karolak
- Routine "organoid_growth" allows to examine the development of multicellular organoids of different morphologies (two distinct organoid shapes shown above): organoid_growth.m & organoid_growth.zip
- This is a companion code that can generate results shown in Fig.1, Fig.3 and Fig.6 from a paper by Karolak, Poonja and Rejniak "Morphophenotypic classification of tumor organoids as an indicator of drug exposure and penetration potential", published by PLoS Computational Biology
Citation: if this code is used, please, cite the following paper:
- A. Karolak, S. Poonja, K.A. Rejniak, (2019) "Morphophenotypic classification of tumor organoids as an indicator of drug exposure and penetration potential", PLoS Computational Biology, 15(7):e1007214, Pubmed#31310602; link to the article
Drug Interstitial Transport Model (2018 Moffitt PhD Program)
This is a part of a graduate lecture in Mathematical Oncology on Tumor Microenvironment, presented at Moffitt in October 2018.
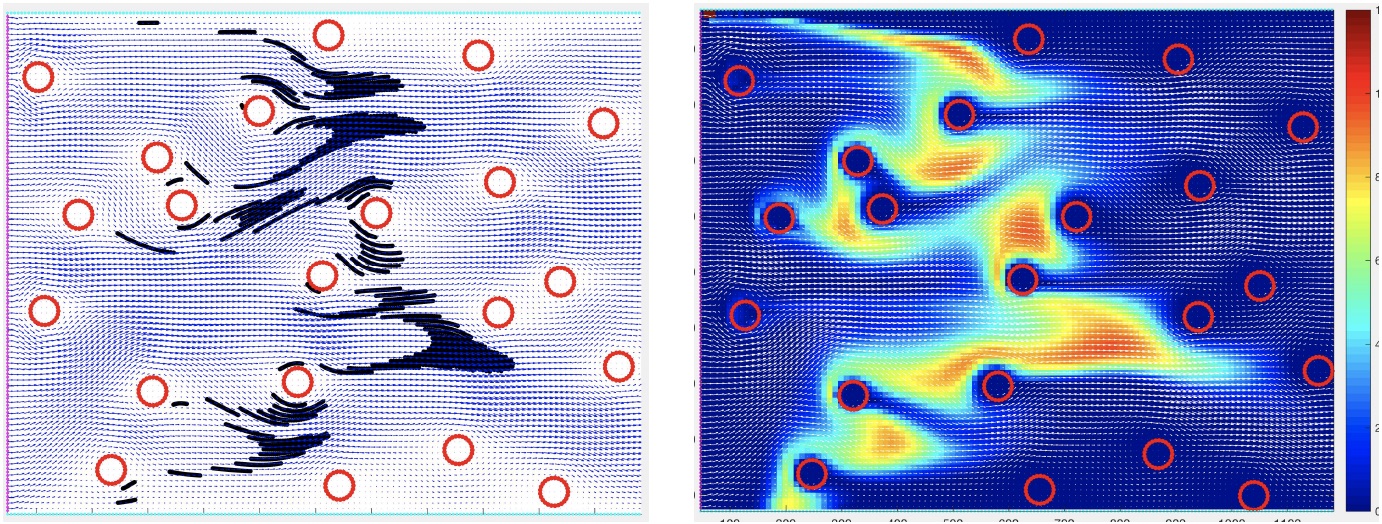
This set of Matlab-based examples to illustrate the interstitial fluid transport of drug molecules in both discrete and continuous description. Created by K.A. Rejniak
- Handouts containing model explanation: Handouts
- Exercise: "interstitial_flow_discrete" illustrates model set-up and advective transport of discrete drug molecules through the tumor interstitium: interstitial_flow_discrete.m
- Exercise: "interstitial_flow_continuous" illustrates advective transport of a concentration of drug molecules through the tumor interstitium: interstitial_flow_continuous.m
- Zip file
Targeted Therapy Model (ACS-IRG & PSOP grants)
This project is supported in part by the by the American Cancer Society Institutional Research Grant ACS-IRG and the NIH-Physical Sciences Oncology Project PSOP grant.
Created by A. Karolak
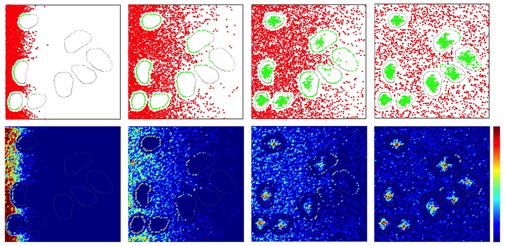
- Routine "tlr2_tumor_code" allows to examine the movement of ligand molecules through the tissue and binding to specific cell receptors; the above images were generated for fast release with moderate binding affinity and slow diffusion and show four differnet snapshots: tlr2_tumor_code.m & tlr2_tumor_code.zip
- This is a companion code generating results shown in Fig.5B top row "Analysis of ligand molecule movement through the tissue space and its role in receptor saturation" from a paper: Karolak et al. "Targeting Ligand Specificity Linked to Tumor Tissue Topological Heterogeneity via Single-Cell Micro-Pharmacological Modeling", Nature Scientific Reports, 8:3638, 2018, link to the paper
Citation: if this code is used, please, cite the following paper:
- A. Karolak, V.C. Estrella, A.S. Huynh, T. Chen, J. Vagner, D.L. Morse, K.A. Rejniak, "Targeting Ligand Specificity Linked to Tumor Tissue Topological Heterogeneity via Single-Cell Micro-Pharmacological Modeling", Nature Scientifric Reports, 8:3638, 2018, link to the paper
Drug Resistance Model (2013 WhAM workshop)
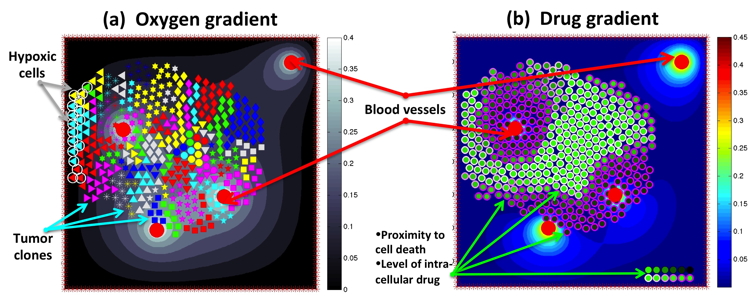
This is a part of a project conducted at the WhAM! Wokshop at the IMA in Minneapolis in September 2013. This set of Matlab-based exercises allows to test different mechanisms of cell resistance (pre-existing and acquired) to anticancer drugs. Created by WhAM! Team: K.A. Rejniak, J. Gevertz, K.A. Norton, A. Volkening, J. Perez-Velazquez and Z. Aminzare. The exercises below guide a user in how to reproduce simulations summarized in Figure 1 of our work published in a book "Applications of Dynamical Systems in Biology and Medicine" that can be find here.
- Handouts containing model description and exercise instructions can be find here: Handouts
- Exercise: "modelWhAM" allows to examine various mechnisms of cell resistance to DNA damaging drug: modelWhAM.m, oxyinit10.txt
- Zip file
Citation: if this code is used, please, cite the following papers:
- J.L. Gevertz, Z. Aminzare, K.A. Norton, J. Pérez-Velázquez, A. Volkening, K.A. Rejniak, "Emergence of Anti-Cancer Drug Resistance: Exploring the Importance of the Microenvironmental Niche via a Spatial Model", Chapter I of the book "Applications of Dynamical Systems in Biology and Medicine", edited by A. Radunskaya and T. Jackson, IMA Volumes in Mathematics and Its Applications, Springer-Verlag, 2015, ISBN 978-1-4939-2781-4; link to the book
- J. Pérez-Velázquez, J.L. Gevertz, A. Karolak, K.A. Rejniak, “Microenvironmental niches and sanctuaries: a route to acquired resistance”, Advances in Experimental Medicine and Biology, 936:149-164, 2016, Pubmed #27739047; PMCID:PMC5113820; link to the article
- J. Pérez-Velázquez, K.A. Rejniak, "Drug-induced resistance in micrometastases: analysis of spatio-temporal cell lineages", Frontiers in Ohysiology, 11:319,2020, doi: 10.3389/fphys.2020.00319 link to publication
2016 HIP-IMO lecture materials:
Created by K.A. Rejniak
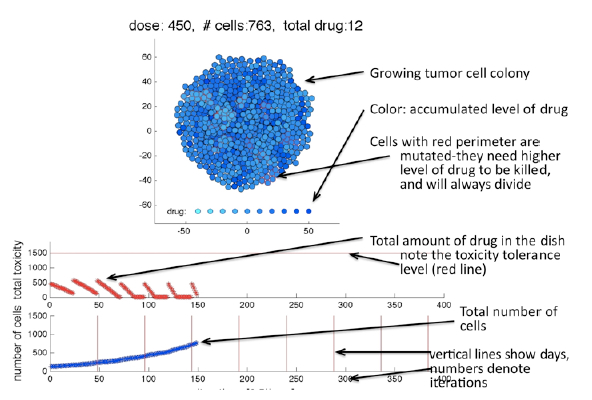
Drug Scheduling Model (2013 Pomona College lecture & 2013 PASI
Workshop & 2016 IMO-HIP lecture)
This is a part of an undergraduate lecture and hands-on exercises session presented at the Pomona Collage in March 2013, and the computer laboratory conducted during the PASI Workshop in Porto Alegre in July 2013.
This set of Matlab-based exercises allows to test different in-vitro scheduling protocols for anticancer drugs. Created by K.A. Rejniak
- Handouts containing model description and exercise instructions can be find here: Handouts
- Exercise: "CellColonyDrug" allows to examine various scheduleing protocols of chemotherapeutic drugs in a model of tumor cell colony: CellColonyDrug.m CellCluster.txt, CellClusterParameters.txt
- Zip file
Clonogenic Assay Model (ICBP grant & Miles for Moffitt Award)
This project is partially supported by the NIH-ICBP grant, and partially by the Miles for Moffitt Milestone Award. Created by MJ. Kim
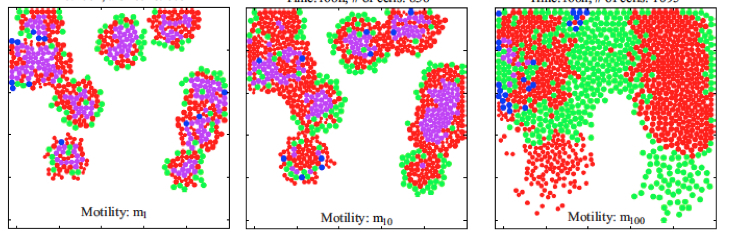
- Routine "CellCulture" allows to examine the impact of cell motility on the formation of cell clusters in 2D cell cultutre model; the above images were generated for motility of 1, 10 or 100, repectively: CellCulture.m CellCulture.zip
- This is a companion code generating results shown in Fig.5 "Collective behavior and spatial organization of cells with different motilities from amanuscript: Kim M, Reed D, Rejniak KA. "The formation of tight tumor clusters affects the efficacy of cell cycle inhibitors:a hybrid model study", published in the Journal of Theoretical Biology. 2014; 352:31-50, open access
Citation: if this code is used, please, cite the following paper:
- M.J. Kim, D. Reed, K.A. Rejniak, "The formation of tight tumor clusters affects the efficacy of cell cycle inhibitors: a hybrid model study", Journal of Theoretical Biology, 352:31-50, 2014; Pubmed ID#24607745, open access
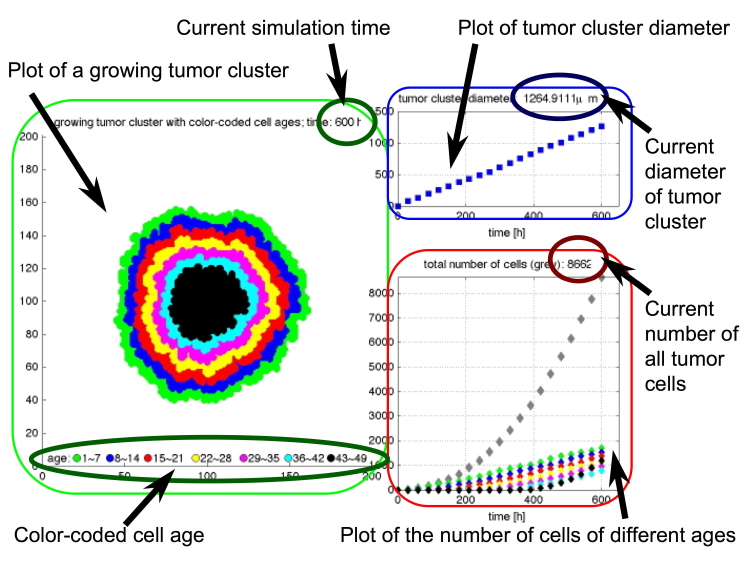
Tumor Growth Model (2010 MBI Cancer Modeling Bootcamp)
This is a part of the Cancer Modeling Bootcamp for the Biologists organized by the MBI in September 2010. This set of Matlab-based exercises shows step-by-step how an agent-based model can be used to investigate growth of a tumor colony. Created by K.A. Rejniak
- Handouts containing model description and exercise instructions can be find here: Handouts
- Exercise 1, "Tumor Growth": allows to examine how growth of the whole tumor cluster depends on individual cell doubling time: Ex1.m
- Exercise 2, "Go or Grow": allows to examine how cell proliferating and moving capabilities contribute to the growth of the whole tumor cluster: Ex2.m, ParamSpace.m
- Exercise 3, "Cell Metabolism", allows to examine how tumor colony will change in response to microenvironmental factors, such as nutrient gradient: Ex3.m
- Exercise 4, "Cell Mutations", allows to examine how tumor cell heterogeneity defined in terms of cell responses to the microenvironmental factors influence the behavior of teh whole tumor - here 6 clones of mutated cells are traced: Ex4.m
- Exercise 5, "All-in-one", this routine combines all features discussed previously and allows to define all properties for a tumor cell base line and the mutants: Ex5.m
- Exercise 6, "3D Tumor Growth", all discussed previously concepts are applied to the 3D cellular automata model: Ex6.m
- Zip file
Deformable Cell Model based on the Immersed Boundary Method
~ IBCell (Institute for Advanced Studies 2002 & Moffitt PhD
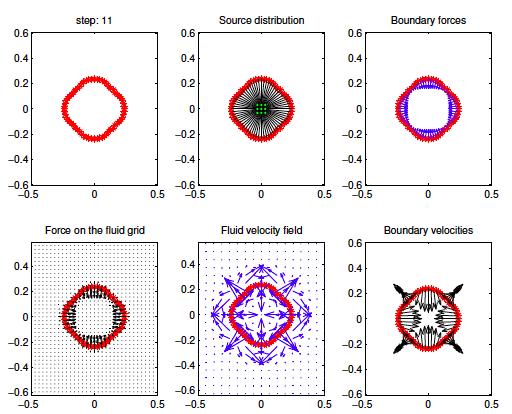
This is a part of a hands-on excercise session prepared for the postgraduate lecture series in the Program for Women in Mathematics on Mathematical Biology held at the Institute for Advanced Study in Princeton, in May 2003. This set of Matlab-base excersices shows step-by-step how the interactions between the fluid and the immersed body are defined in the immersed boundary method -- here applied to a growing cell of either circular or squared shape with a pseudo-skeleton imposed by spring forces. Created by K.A. Rejniak
- Routine "CellGrowth" illustrates the way the immersed boundary method is implemented (each step is shown above):
- a cell is composed from boundary points that form either a circle or a square (row 1, column 1);
- the adjacent boundary points are connectd by springs to form cell membrane; additional springs are defined to form a pseudo-cytoskeleton (row 1, column 2);
- springs that are not in a resting distance exert forces on the boundary points (row 1, column 3);
- these forses are transmitted to the fluid (row 2, colum 1);
- the fluid flow arising form the centrally located fluid source and all boundary and cytoskeletal forces is computed (row 2, column 2);
- fluid flow velocities are extrapolate to all boundary points (raw 2, column 3).
- CellGrowth.m
- Zip file
Drug Penetration Model (PSOC Network & 2013 PASI Workshop)
This is a part of the computer laboratory conducted during the PASI Workshop in Porto Alegre in July 2013. Developement of this model was partially funded by the PSOC Network. This set of Matlab-based exercises allows to test different in-vivo scheduling protocols for anticancer drugs. Created by K.A. Rejniak under construction ... please, contact me if you are interested